Adapting protein aligner parameter choices to match varying mutation rates
Dan DeBlasio and John KececiogluE-mail question/comments: deblasio@cs.arizona.edu
About
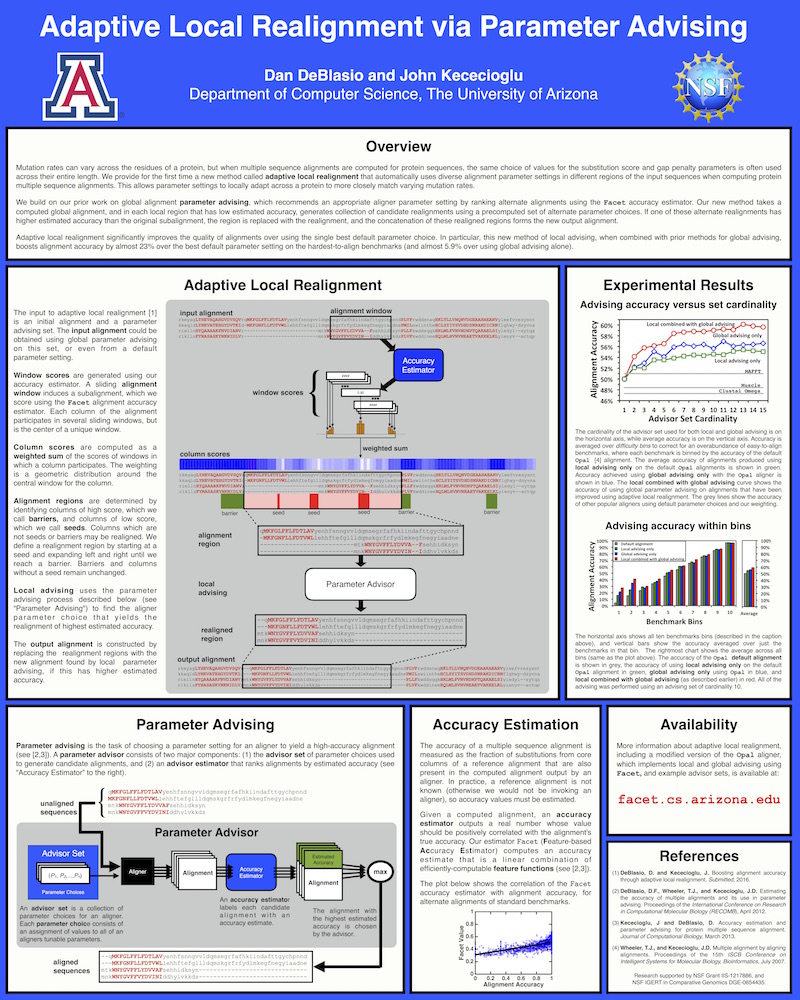
Mutation rates may vary along the length of a protein, thus choosing a single multiple sequence alignment parameter choice may not be appropriate. To overcome this we have developed a method called Adaptive Local Realignment which uses parameter advising to choose alternate alignment parameters choices as the underlying mutation rate changes. To accomplish this we first use Facet to compute column scores for an alignment (by estimating accuracy in sliding windows across the alignment) and identifying columns that are very low quality (seeds) and very high quality (barriers). We can then extend the seeds to the left and right until they reach a barrier to create realignment regions. For these regions we use parameter advising to select a parameter choice that generates a more accurate sub-alignment.
Currently adaptive local realignment is integrated into the development version of the Opal aligner which is available on GutHub. A related publication that explains the details, as well as a conference poster will be available soon.
Publications
Please cite:
Boosting alignment accuracy through adaptive local realignment
Dan DeBlasio and John Kececioglu
Proceedings of the 21st
Conference on Research in Computational Molecular Biology (RECOMB) 2017,
Springer-Verlag Lecture Notes in Computer Science 10229, 1-17, 2017.
doi:10.1007/978-3-319-56970-3_1
(pdf)
(talk)
Poster
This poster was presented at ISMB 2016 in Orlando, FL, USA and descries adaptive local realignment.
Slides from my talk is ISMB 2016 can be found here. |
Download
Adaptive local realignment is included in the newest version of the Opal aligner. The Opal distribution includes the Opal and Facet software as a .jar file as well as a driver and the scripts needed to predict secondary structure. Advisor sets can be downloaded below.
Opal v3.1.b0 (tgz) (14 Jun 2016)
The newest version of the Opal development trunk contains adaptive local realignment. You can find the development version of Opal from the Opal GitHub (http://git.io/Opal)
Note this application requires a working copy of PSIPRED as well as BLAST. PSIPRED v3.2 can be downloaded here: (link).
Advisor Sets
Oracle advisor sets
This file contains the parameter sets used for analysis for the Facet Kececioglu and DeBlasio 2013. There is a README in the attached file as well that explains the format. These features were used to generate the alternate alignments of the benchmarks used for evaluation. (tgz)
Supplemental Data
Benchmarks
The protein benchmarks used for testing Facet are a combination of the BENCH (link) benchmark set or Edgar and the PALI (link) benchmark set. Attached is a tar file wth both the input sequence sets as well as the reference alignment annotated with the core columns in capital letters. (tgz). The PSIPRED structure predictions can also be downloaded here: (tgz)
Acknowledgements
Research supported by the NSF IGERT Grant in Comparative Genomics DGE-0654435 and NSF GrantIIS-1217886.
